FCS Files¶
Adding FCS Files to an Experiment¶
Uploading¶
Howto
- Drag and drop folders or files into your experiment. Uploads will be tracked in the upper-right corner. You can navigate to other pages within CellEngine during the upload.
If you attempt to upload FCS files to an experiment that already has files with the same filenames uploaded, you will be given the option of skipping the duplicate files and only uploading the new files.
Importing FCS Files from Other Experiments¶
Importing copies an FCS file from another experiment into the current experiment.
Howto
- In the FCS file list on the experiment summary page, click import FCS file.
- Select the experiment from which you want to import the file.
- Select the FCS file that you want to import.
- Click import.
Importing and Exporting FCS Files from and to S3-Compatible Services¶
FCS files can be imported from and exported to S3-compatible services (AWS S3, Google Cloud Storage, etc.) using the CellEngine API.
Changing FCS File Names¶
In some cases, it is helpful to change the names of FCS files after they have been uploaded to CellEngine. By default, the ability to rename files is locked to prevent any accidental changes.
Howto
- Go to the Annotations page.
-
Click the option Change filenames in the toolbar.

-
Double click on the name of any FCS file and type your corrections.
To protect your changes, the Change filenames option will be disabled once you leave the Annotations page.
FCS File Compatibility¶
CellEngine has industry-leading data file compatibility, with support for FCS2.0, FSC3.0 and FCS3.1. We continually validate with data files from:
- Acea Bio: NovoCyte
- Apogee Flow: Micro, Universal, Auto40
- Attune: Attune (original and new)
- Beckman Coulter: Cyan, Epics XL-MCL, CytoFLEX, Gallios, Navios, MoFlo Astrios, MoFlo XDP, Cytomics FC 500
- Beckton Dickinson (BD): Accuri C6, FACSAria, FACSCalibur, FACSCantoII, FACSort, FACSVerse, LSRFortessa, LSRII, Symphony X50, FACSLyric
- Cytek: Aurora, Northern Lights, dxP10, dxP8, xP5
- Standard Biotools (formerly Fluidigm): CyTOF1, CyTOF2, Helios
- Luminex: Amnis ImageStream Mk II
- Millipore: easyCyte 6HT, Guava
- Miltenyi Biotec: MACSQuant
- Partec: CyFlo Cube 6, PAS
- Propel Labs: YETI/ZE5
- Sartorius: Intellicyt iQue 3
- Sony: iCyt Eclipse, SA3800 Spectral Analyzer
- Stratedigm: S1400
CellEngine tolerates invalid FCS files in most cases, and has the ability to correct invalid spill strings (compensation matrices), unescaped delimiters, empty keywords, incorrect segment coordinates and other violations.
CellEngine also generally applies the same adjustments that the instrument manufacturers apply in their own software, which includes vendor-specific scaling and transformations not captured in the FCS specification.
Viewing FCS File Metadata¶
You can view meta-information about FCS files, such as FCS header values, laser delays and PMT voltages, by clicking on the three-dot menu for a file in the list of FCS files on the experiment summary page, and then selecting view details.
For CyTOF Helios files that have only been processed with the Fluidigm Helios software, this dialog also displays the last tuning results and other instrument performance information.
Tip
FCS header values can also be added to the annotations for a file, allowing for further analysis using header parameters.
Concatenating FCS Files¶
Concatenating merges two or more files into one.
Howto
- In the FCS file list on either the experiment summary page or the annotation page, select the files that you want to concatenate.
- In the menu bar above the file list, select concatenate files. A dialog will open.
- Select whether or not to add a file number column to the output file.
- Select whether or not to delete the input files after successfully concatenating.
- Click concatenate. The concatenated file will be added to your experiment.
You can elect to add a file number column (channel) to the output file so that you can see which file each event (cell) came from. The values in this column have a uniform random spread (±0.25 of the integer value) to ease visualization. While this column can be useful for analysis, it will cause your experiment to have FCS files with different panels unless you delete all FCS files that have not been concatenated.
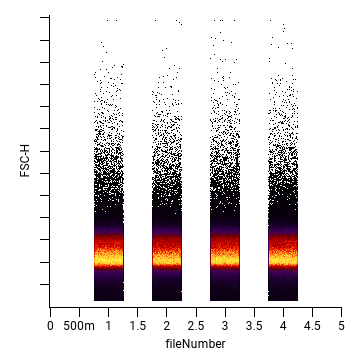
During concatenation, any FCS header parameters that do not match between files will be removed, with some special exceptions:
$BTIM(clock time at beginning of acquisition) and$DATEwill be set to the earliest value among the input files.$ETIM(clock time at end of acquisition) will be set to the latest value among the input files.$PnR(range for parametern) will be set to the highest value among the input files.
All channels present in the first selected file must also be present in the other selected files.
Control Files¶
Files set as “control files” in the annotation page will only be visible in the compensation editor, the experiment summary and the annotations page. This lets you exclude compensation controls, calibration beads and other files that you don’t want to see in your analysis.